Differences between initiation of prokaryotes and eukaryotes: In translation process, main difference between eukaryotes and prokaryotes is in the initiation process itself. Some major differences between Eukaryotic initiation and prokaryotic initiation are as follows. In Eukaryotes, there is only one start codon for Eukaryotes AUG and it codes for methionine and not N-formyl-methionine. Eukaryotic cells need more initiation factors than prokaryotes. Eukaryotic cells require 12 initiation factors. In Eukaryotes, process of association of mRNA with smaller subunit (40s) is more complex than prokaryotes. 40s first subunit identifies 5’ methylated cap of m-RNA and then there is scanning process involved wherein initiation codon is recognized. This recognition is aided by ATP dependant helicases that hydrolyse ATP. This recognition of initiation codon is also aided by Kozak sequences 5’-ACCAUGG-3’ similar to Shine-dalgarno sequence in prokaryotes Figure 30.8.
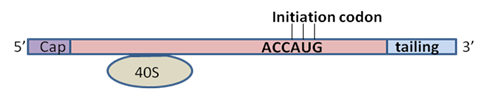
Figure 30.8 : Ribosome binding site in eukaryotes
Important thing to notice that in prokaryotes there is no scanning process, 16s subunit directly binds to region so that initiation codons is in P-site whereas in eukaryotes there is proper scanning process.
However, at first, initiation factor eIF3 and eIF2 binds 40s subunit where the former factor prevents premature association of 60s subunit with 40s and later factor which is GTP binding trimeric protein binds to tRNAMet (ternary complex= eIF2+GTP+tRNAMet ) . Ternary complex and eIF3 binds to 40s subunit forming 43s initiation complex. All this happens without the presence of mRNA.
Then mRNA binding to 40s takes place with the help of initiation factor eIF4F. It is a heterotrimer which consists of eIF4G, eIF4E, and eIF4A. eIF4E binds to 5’ cap and eIF4A acts as helicase to unwind any secondary structure at 5’ end. eIF4G interacts with poly (A) tail via poly (A) binding protein associated (PBP). It implies that there is some sort of circular organization during protein synthesis.