4.10.4. 4. DNA MAJOR GROOVES AND MINOR GROOVES
More recent studies used X-ray crystallography and NMR to confirm the original structure by Watson and Crick. Right handed helix- winds about the same direction in which fingers of a right hand curl when the thumb is pointing upward.
Helical axis passes through base pairs and
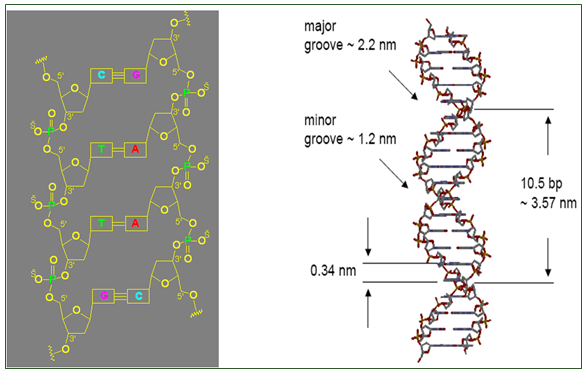
- Contains a minor and a major groove that wind about the outside of the helix
- Helical structure repeats every 34 A
- Major and minor groove are the two surfaces that wind about the outside of the helix. They are formed by the edges of the stacked bases. These grooves are distinct because they have different H-bonding patterns and different size.
- Major and major groove of DNA contain sequence-dependent patterns of H-bond donors and acceptors.
- Sequence-dependent duplex structure (A, B, Z, bent DNA).
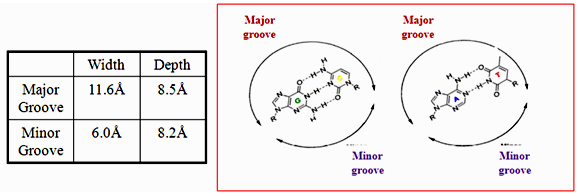
Figure 4.1: Groove dimensions and the WC base pairs.
- The grooves are formed by the edges of stacked bases and have different sizes because deoxyribose residues are attached asymmetrically (not 180 degrees). The minor groove is the one in which C-1’ –helix axis’c1’ angle is less than 180 degrees (recall that the helix axis passes trough the middle of each base pair in B-DNA) and the major groove is on the opposite side of each base pair.
- A/T sequences have a higher negative potential while G/C sequences have higher positive potential
- H-bonding to base edges in the minor groove can be used to distinguish between A:T and G:C base pairs
- A:T is distinguished from a T:A base pair through indirect read out
- DNA binding drugs are generally flat and small, which makes them fit well into the minor groove.
- Groove width varies with sequence; A/T rich tracks tend to make the groove width narrower.
- The convex shape of the minor groove floor complements the typical shapes of minor groove binding drugs
- A/T sequences result in a smooth convex curve whereas G/C sequences have “little” bumps due to the 2-amino groups of guanine.
4.10.4. 5. Sugar-Phosphate Chain Conformations
- Double-stranded DNA has limited structural complexity compared to proteins (only 4 nucleotides vs. 20 amino acids)
- Limited secondary structures, no tertiary or quaternary structures.
- RNA has some well-defined tertiary structure.
- Conformation of a nucleotide is specified by 6 torsion angles of the sugar phosphate backbone and the torsion angle that describes the orientation of the base about the glycosidic bond (7 totals).
- Despite 7 degrees of freedom per nucleotide, they have restricted conformational freedom.
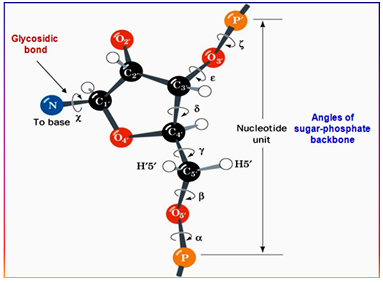
Figure 4.2: The conformation of a nucleotide unit defined by the seven torsion angles.