Funcional structure of a gene:
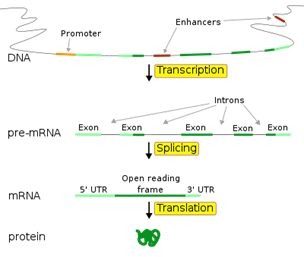
Fig 11 . Diagram of the "typical" eukaryotic protein-coding gene. Promoters andenhancers determine what portions of the DNA will be transcribed into theprecursor mRNA (pre-mRNA). The pre-mRNA is then spliced into messenger RNA(mRNA) which is later translated into protein.
- All genes have regulatory regions in addition to regions that explicitly code for a protein or RNA product. A regulatory region shared by almost all genes is known as the promoter, which provides a position that is recognized by the transcription machinery when a gene is about to be transcribed and expressed. A gene can have more than one promoter, resulting in RNAs that differ in how far they extend in the 5' end. Although promoter regions have a consensus sequence that is the most common sequence at this position, some genes have "strong" promoters that bind the transcription machinery well, and others have "weak" promoters that bind poorly. These weak promoters usually permit a lower rate of transcription than the strong promoters, because the transcription machinery binds to them and initiates transcription less frequently. Other possible regulatory regions include enhancers, which can compensate for a weak promoter. Most regulatory regions are "upstream"—that is, before or toward the 5' end of the transcription initiation site. Eukaryotic promoter regions are much more complex and difficult to identify than prokaryotic promoters.
- Many prokaryotic genes are organized into operons, or groups of genes whose products have related functions and which are transcribed as a unit. By contrast, eukaryotic genes are transcribed only one at a time, but may include long stretches of DNA called introns which are transcribed but never translated into protein (they are spliced out before translation). Splicing can also occur in prokaryotic genes, but is less common than in eukaryotes.
Of the two nucleotide strands of the DNA of a gene, only one strand contains coded genetic information and directs synthesis of RNA via a cell process called DNA transcription. This strand is called the template strand . During DNA transcription, only one strand of the double-helix of a gene, the so-called sense or template strand , serves as the so-called template for DNA transcription to form the new RNA molecule. While during DNA replication certain enzymes called helicases help to unwind and open the DNA double helix at the ori, it is the RNA polymerase enzyme which unwinds DNA during DNA transcription to create single stranded DNA regions.The distinct places along the DNA strand where transcription begins are called transcription start sites. - at the transcription start site, a complex protein cluster, the so-called transcriptome , assembles itself; in eukaryotic cells, the transcriptome is comprised of many protein components. Along these unwind and single-stranded DNA regions, new nucleotides are paired according to the Watson-Crick base-pairing rule (--> A with T; and G with C). The polymerization of the new nucleotides during the process of DNA transcription
is catalyzed by an enzyme called RNA polymerase (RNA POL). The RNA polymerase recognizes certain structures on the DNA strand, called promoter regions, or promoters. Promoters are sequences of DNA which are usually located upstream from the actual coding or transcribed region of a gene. Different genes have promoters with different DNA sequences. In E.coli the promoter has two different functions which relate to two different regions within the promoter, each with a unique consensus sequence:
1. RNA polymerase recognition site
- Mediated by the RNA polymerase recognition site, with the consensus sequence 5'- TTGACA -3' on the non-template strand of the gene
- The site of the initial association of the RNA polymerase enzyme with the gene
- Located about 35bp (-35 region) away from the transcription start site
2. RNA polymerase binding site
- this DNA sequence is also known as the "Pribnow box"
- centered at the -10bp region of the gene
- this is the region where the RNA polymerase enzyme starts to localized unwinding of the
DNA double strand of the gene
- both gene regions tells the ribosome where to dock on and where to start the DNA transcription process on a gene
- promoter regions often contain important regulatory sites , such as the operator region or the
CAP binding site (see bacterial gene regulation!), which allow the fine tuning of DNA transcription depending on different environmental conditions.
Another important stretch of DNA on a gene is the so-called leader sequence .
- this gene region located downstream of the promoter region is transcribed first into mRNA, but is does NOT contain a translated codon;
- it is a non-translated DNA sequence with two known functions:
1. it is important for initiation of protein translation at the small subunit of the ribosome
- this happens with the help of the Shine-Dalgarno consensus sequence , (5'- AGGA -3'),
which is complementary to a sequence on the 16S rRNA transcript
2. it is involved in the regulation of DNA transcription , e.g. during attenuation.
Following the non-transcribed leader sequence of a gene follows the coding region .
- this code carrying region directs the actual synthesis of the polypeptide chain at the ribosome
- a coding sequence of a gene usually begins with the consensus sequence 5'- TAC -5';
- this sequence after transcription by the RNA polymerase into the RNA translation initiation codon 5'- AUG -3' is translated by the ribosome into the amino acid N-formylmethionine;
- the remainder of the coding region of a gene consists of a unique sequence of triplet codons, e.g. UCG, that specifies the sequence of amino acids for that particular gene;
Another important sequence or region on a typical gene is the terminator sequence.
- these gene region located at the end of a gene sequence is important to enforce the termination of DNA transcription by stopping the RNA polymerase;
- it often after a non-transcribed so-called trailer sequence, which is important for the proper
expression of the coding region of a gene.
In order to begin gene transcription, the RNA polymerase enzyme further requires a number of helper proteins, the so-called transcription factors (TFs)
- important bacterial transcription factors Sigma and Rho;
- important eukaryotic transcription factors are myc, AP-1, TFIIA, TFIIB, etc.;
- the RNA polymerase plus the transcription factors recognize and bind to the so-called promoter region ( TATA box ), which is located shortly before the beginning of a typical gene
REFERENCES:
Text Books:
1. Jeffery C. Pommerville. Alcamo's Fundamentals of Microbiology (Tenth Edition). Jones and Bartlett Student edition.
2. Gerard J. Tortora, Berdell R. Funke, Christine L. Case. Pearson - Microbiology: An Introduction. Benjamin Cummings.
3. J. Krebs, E.S. Goldstein, Stephen T. Kilpatrick. Lewin's Genes X. Jones and Bartlett Publishers.
Reference Books:
1. Lansing M. Prescott, John P. Harley and Donald A. Klein. Microbiology. Mc Graw Hill companies.