Genetic code is degenerate. Since there are more codons than the amino acids, more than one codon may specify the same amino acid. Such different codons that specify the same amino acid are called as synonymous codons . e.g., UUU = UUC = phenylalanine.
Genetic code has start/stop signals. Some codons are specially meant for initiation and termination of protein synthesis. e.g.: AUG codes for methionine, serves as initiation codon in eukaryotes and GUG in case of prokaryotes. Three codons UAG (amber), UAA (ochre), UGA (opal) are called as termination codons, because they terminate protein synthesis. Earlier, they were called as nonsense codons, because they do not code for any amino acid, but, since they are involved in termination of protein synthesis, they are called as termination codons. The initiation and termination codons are known as signals and this phenomenon is known as punctuation.
Genetic code is polar. It means that the genetic code has a fixed start and termination ends, and is always read in a fixed direction, i.e. in 5'→ 3' direction and the polypeptide chain is synthesized in N→C direction i.e., from amino group (NH 2) to carboxylic group (COOH).
Genetic code is universal. The same genetic code is applicable to all organisms, from bacteria to man, i.e.; the codons have the same meaning in all the organisms. e.g., UUU = phenylalanine in bacteria, mouse, man and tobacco. The universality of the genetic code, however, does not mean that DNA base ratios must be similar in different species for genes specifying similar proteins. The fact that the code is degenerate enables many bases to be changed by mutation in a sequence of mRNA, but this mRNA could still produce the same amino acid sequence.
In 1979 investigators began started DNA sequencing of mitochondrial DNA in humans, cattle and mice. During their experiments, they were surprised to learn that the genetic code used by the mitochondrial DNA was not the same as the universal genetic code. e.g. UGA, which is a non-sense codon, but it codes for tryptophan in mtDNA, AGG which codes for arginine is a non-sense codon in mtDNA. So, extra chromosomal DNA such as mtDNA and ctDNA do not come under the universality of the genetic code.
Wobble Hypothesis. Out of the 64 codons, three are involved in termination process. So, there are only 61 codons specifying the amino acids, and the cell should have 61 different types of tRNAs, each having a different anticodon for the recognition of codons. However, the actual number of tRNA is found to be much less than 61. This means that the anticodons of tRNA read more than one codon on the mRNA. Crick (1966) proposed a hypothesis to explain the degeneracy of the genetic code; the hypothesis is known as Wobble hypothesis.
According to this hypothesis, the major degeneracy occurs at the third position, i.e., the third codon is not important in base pairing, and the actual pairing occurs only in the first two codon-anticodon pairs. The base at the 5′ end of the anticodon and the base at the 3′ end of the codon form hydrogen bonds without any specificity. The third base is called as the wobble base . This wobble base of codon lacks specificity and the base in the first position of the anticodon is usually abnormal e.g., inosine, tyrosine, etc. These abnormal bases are able to pair up with more than one nitrogen base at the same position e.g., inosine (I) can pair up with A, C and U. The pairing between unusual base of tRNA and wobble base of mRNA is called wobble pairing.
Gene structure:
A gene is a linear sequence of nucleotides or codons (= follow-up of three nucleotides) with a fixed start and end point. In its most simplified version one gene codes the information for one enzyme (--> "one gene - one enzyme hypothesis"). This hypothesis has been modified over the years to the "one gene - one polypeptide" hypothesis, which holds true for most bacterial genes. Many enzymes and proteins for which genes carry the code are made up from more than one polypeptide requiring more than one gene. "The DNA segment which codes for a single polypeptide sequence is often referred to as a cistron."
Most genes consist of discrete sequences of codons that are "read" (transcribed) on only one way to generate one translated polypeptide sequence. The code is NOT overlapping and there is only one defined start point with one reading frame, i.e. the way in which the DNA nucleotides are arranged in codons. While each gene harbors the genetic information of the DNA strand and typically codes for a specific protein or enzyme, viral, prokaryotic and eukaryotic genes structures differ significantly. In contrary to bacterial genes, genes in eukaryotic life forms are more spaced apart and also are organized in so-called exon-intron structure ;- exons are coding segments of a genes, while introns are non-coding gene regions which play a role in splicing and gene regulation; - genetic information of cistrons in prokaryotes is usually continuous and introns rarely occur; moreover, bacterial genes are often organized in so-called operons ;- transcription of bacterial operons generates polygenic or polycistronic mRNA , i.e. which is RNA that is translated into more than one polypeptide.
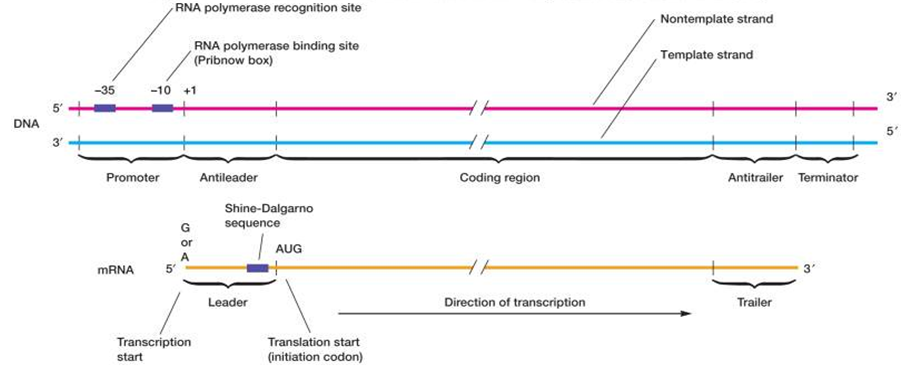
Fig. 10. A bacterial structural gene.