Biosynthesis of the sugar moieties of calicheamicin ![]() : Biosynthesis of monosaccharides-The Aryl Tetrasaccharide of Calicheamicin
: Biosynthesis of monosaccharides-The Aryl Tetrasaccharide of Calicheamicin ![]() : Calichiamicin
: Calichiamicin![]() contains four highly modified monosaccharides. Two of the monosaccharides are linked via an unusual hydroxylamino glycosidic linkage. All four sugars are deoxysugars that exhibit greater hydrophobicity compared to common sugars. The deoxysugars along with the linked orsellenic acid group is giving the DNA binding specificity of calicheamicin
contains four highly modified monosaccharides. Two of the monosaccharides are linked via an unusual hydroxylamino glycosidic linkage. All four sugars are deoxysugars that exhibit greater hydrophobicity compared to common sugars. The deoxysugars along with the linked orsellenic acid group is giving the DNA binding specificity of calicheamicin![]() . The biosynthesis of the four monosaccharides starts with the conversion of glucose-1-phosphate to NDP-glucose which is similar to the biosynthetic pathway of sugar for C-1027, neocarzinostatin, maduropeptin and other glycosides. Analysis of gene cluster of Calichiamicin
. The biosynthesis of the four monosaccharides starts with the conversion of glucose-1-phosphate to NDP-glucose which is similar to the biosynthetic pathway of sugar for C-1027, neocarzinostatin, maduropeptin and other glycosides. Analysis of gene cluster of Calichiamicin ![]() reveals that CalS7 catalyzes the formation of NDP-glucose in the early stage of synthesis of sugars A–D. As discussed below, the pathways for the four sugars would diverge after the formation of NDP-glucose. The 4-hydroxyamino-6-deoxy-a-D-glucose (sugar A) is directly tethered to the enediyne core. The deoxygenation at C6 position and installation of the amino group are followed similar routes that involve the NDP-4-keto-6-deoxy-a-glucose intermediate in case of C-1027 (Scheme 20).
reveals that CalS7 catalyzes the formation of NDP-glucose in the early stage of synthesis of sugars A–D. As discussed below, the pathways for the four sugars would diverge after the formation of NDP-glucose. The 4-hydroxyamino-6-deoxy-a-D-glucose (sugar A) is directly tethered to the enediyne core. The deoxygenation at C6 position and installation of the amino group are followed similar routes that involve the NDP-4-keto-6-deoxy-a-glucose intermediate in case of C-1027 (Scheme 20).
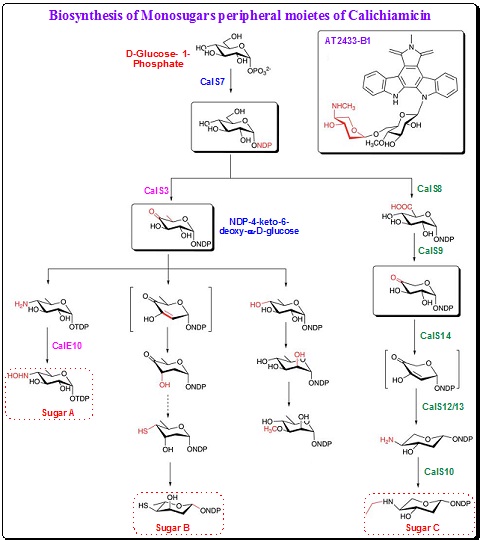 |
Scheme 20. Biosynthetic pathways for the monosaccharides of calicheamicin |
The hydroxylamino glycosidic bond between sugar A and sugar B is very crucial for maintaining the correct conformation and rigidity of the oligosaccharide for effective binding to the minor groove of DNA. A novel aminosugar oxidase, the N-oxidase CalE10, enzyme catalyzes the hydroxylation of the amine group. CalE10 is a heme protein that exhibits stringent regiospecificity with limited overoxidation in vitro.
Calicheamicin ![]() contains a 4-thio-2,4,6-trideoxy-D-glucose (sugar B) that is connected to the orsellenic acid moiety through a thioester linkage. The sulfur atom of sugar B contributes to the binding mode of calicheamicin
contains a 4-thio-2,4,6-trideoxy-D-glucose (sugar B) that is connected to the orsellenic acid moiety through a thioester linkage. The sulfur atom of sugar B contributes to the binding mode of calicheamicin ![]() by forming hydrogen bonds with an exposed amino proton of guanine in the minor groove of DNA. However the mechanism of installation of sulfur atom is not known. The closely related esperamicin A1 also contains a similar thiosugar. From the isotope feeding experiment, it is known that the sulfur is derived from L-methionine or L-cysteine.
by forming hydrogen bonds with an exposed amino proton of guanine in the minor groove of DNA. However the mechanism of installation of sulfur atom is not known. The closely related esperamicin A1 also contains a similar thiosugar. From the isotope feeding experiment, it is known that the sulfur is derived from L-methionine or L-cysteine.
The enzymes involved in the biosynthesis of methylated 3-methoxy-L-rhamnose (sugar C) synthesis remains unclear. However, it is believed that the biosynthesis follow a common pathway for rhamnose synthesis with the deoxygenation at C6 facilitated by the formation of the NDP-4-keto-6-deoxy-a-D-glucose intermediate (Scheme 20). A putative epimerase and a methyltransferase most probably catalyze the epimerization at C2 and methylation at C3 respectively.
Aminodideoxypentose (4-amino-3-O-methoxy-2,4,5-trideoxypentose, sugar D) present in calicheamicin ![]() is similar to that present in esperamicin A1 and in indolocarbazole antitumor antibiotic AT2433. Comparison of the calicheamicin and AT2433 gene clusters indicated the presence of a set of common genes that are expectedly involved in the biosynthesis of the 4-amino-3-O-methoxy-2,4,5-trideoxypentose.
is similar to that present in esperamicin A1 and in indolocarbazole antitumor antibiotic AT2433. Comparison of the calicheamicin and AT2433 gene clusters indicated the presence of a set of common genes that are expectedly involved in the biosynthesis of the 4-amino-3-O-methoxy-2,4,5-trideoxypentose.
It is established that the synthesis of the modified pentose proceeds via a TDP-sugar pathway. A decarboxylation step is involved at C6 position with the help of a UDP-glucuronate decarboxylase-like protein (CalS9). The deoxygenation of C2 and installation of amino group at C4 follow as per the mechanisms shown in Scheme 20.
Assembly of calicheamicin ![]() by glycosyltransferases: The orsellenic acid and sugar moieties are assembled onto the enediyne aglycone in the final stages of calicheamicin biosynthesis. A set of O-glycosyltransferases are utilized to catalyze the transfer of the sugars from the nucleotide diphosphate sugar donors to the aglycone acceptor. Glycosyltransferases CalG3 and CalG2 catalyze the first two glycosylation steps in a sequential fashion (Figure 21). The crystal structure of CalG3 revealed a typical UDP-glycosyltransferase/glycogen phosphorylase fold found in many glycosyltransferases. It is also known that CalG3 catalyzes the formation of the unusual hydroxylamino glycosidic bond by using TDP-4,6-dideoxy-a-D-glucose as a substrate surrogate. The other two putative glycosyltransferases (CalG1 and CalG4) encoded by the gene clusters are the rhamnosyltransferase and aminopentosyltransferase which are responsible for the installation of sugars C and D respectively.
by glycosyltransferases: The orsellenic acid and sugar moieties are assembled onto the enediyne aglycone in the final stages of calicheamicin biosynthesis. A set of O-glycosyltransferases are utilized to catalyze the transfer of the sugars from the nucleotide diphosphate sugar donors to the aglycone acceptor. Glycosyltransferases CalG3 and CalG2 catalyze the first two glycosylation steps in a sequential fashion (Figure 21). The crystal structure of CalG3 revealed a typical UDP-glycosyltransferase/glycogen phosphorylase fold found in many glycosyltransferases. It is also known that CalG3 catalyzes the formation of the unusual hydroxylamino glycosidic bond by using TDP-4,6-dideoxy-a-D-glucose as a substrate surrogate. The other two putative glycosyltransferases (CalG1 and CalG4) encoded by the gene clusters are the rhamnosyltransferase and aminopentosyltransferase which are responsible for the installation of sugars C and D respectively.
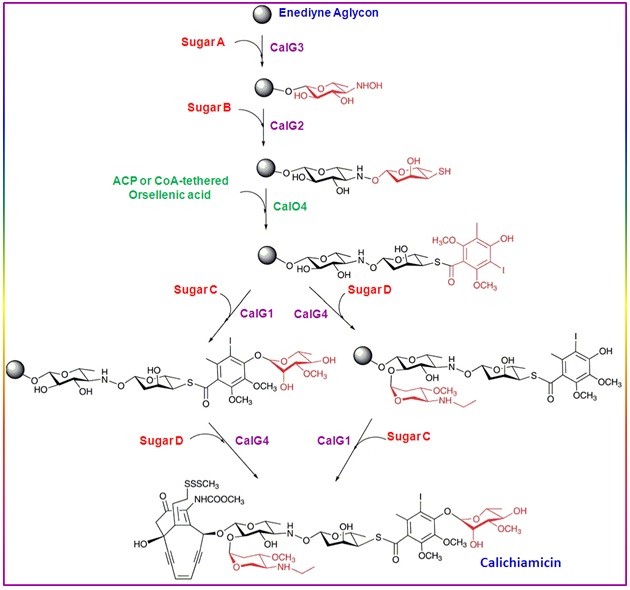 |
Figure 21. Assembly of calicheamicin |