2.5 Biosynthesis of 10-Membered Ring Enediynes
Peripheral moieties of the 10-membered enediynes: Based on the structure and folding pattern of the enediyne core the 10-membered enediyne chromophores are further divided into two sub group--(i) calicheamicin-like and (ii) dynemicin-like enediynes. Calicheamicin-like enediynes include (a) esperamicins, (b) namenamicin and (c) shishijimicins. These are decorated with a variety of sugar and aromatic peripheral moieties. In comparison, the dynemicin-like chromophores include (a) dynemicin and (b) uncialamycin. These are characterized by the presence of an anthraquinone peripheral moiety. The calicheamicin and dynemicin gene clusters have been sequenced and partially annotated. Studies revealed the presence of enzymes involved in the biosynthesis of the peripheral moieties of calicheamicin ![]() are the sugar-modifying enzymes and glycotransferases.
are the sugar-modifying enzymes and glycotransferases.
2.5.1 Biosynthesis of Calicheamicin ![]()
Calicheamicins were isolated from Micromonospora echinospora spp. calichensis from caliche soils. Calicheamicins represent a subfamily of enediyne chromophores and their structure, mode of action and pharmacological properties have been examined rigorously. Calicheamicin ![]() , the best characterized member of the calicheamicins, contains a complex aryltetrasaccharide moiety that confers the chromophore its DNA-binding specificity. The Building blocks of Calicheamicin
, the best characterized member of the calicheamicins, contains a complex aryltetrasaccharide moiety that confers the chromophore its DNA-binding specificity. The Building blocks of Calicheamicin ![]() are (a) enediyne core, (b) aryltetrasaccharide that is composed of an iodized orsellenic acid moiety and four monosaccharides. The aryltetrasaccharide unit is characterized by an unusual thiosugar and a hydroxylamino glycosidic linkage (Figure 19).
are (a) enediyne core, (b) aryltetrasaccharide that is composed of an iodized orsellenic acid moiety and four monosaccharides. The aryltetrasaccharide unit is characterized by an unusual thiosugar and a hydroxylamino glycosidic linkage (Figure 19).
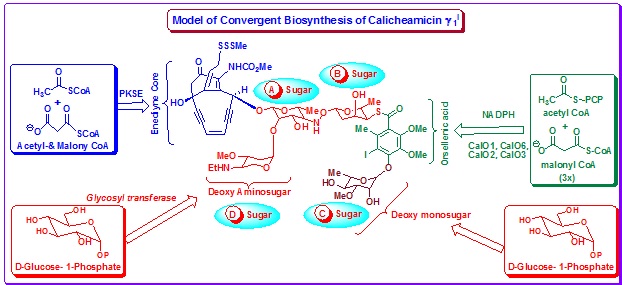 |
Figure 19. Building blocks and model of biosynthesis of Calicheamicin |
Biosynthesis of Orsellenic acid of aryltetrasaccharide peripheral unit of Calicheamicin ![]() : In addition to calE8, a second iterative PKS (calO5) gene is present in the Calicheamicin
: In addition to calE8, a second iterative PKS (calO5) gene is present in the Calicheamicin ![]() cluster. The predicted gene product (CalO5) shares significant sequence homology with AviM, the iterative PKS for the biosynthesis of the oligosaccharide antibiotics avilamycins produced by Streptomyces viridochromogenes. Notably, AviM is also the first type I polyketide synthase isolated from bacteria to produce an aromatic compound. The fact that avilamycins and the related evernimicin also contain a chlorinated orsellenic acid group hints that CalO5 could be involved in the synthesis of the iodized orsellenic moiety in calicheamicin.
cluster. The predicted gene product (CalO5) shares significant sequence homology with AviM, the iterative PKS for the biosynthesis of the oligosaccharide antibiotics avilamycins produced by Streptomyces viridochromogenes. Notably, AviM is also the first type I polyketide synthase isolated from bacteria to produce an aromatic compound. The fact that avilamycins and the related evernimicin also contain a chlorinated orsellenic acid group hints that CalO5 could be involved in the synthesis of the iodized orsellenic moiety in calicheamicin.
Sequence analysis suggested that CalO5 contains at least four domains (KS, AT, DH and ACP) with a ‘core’ domain apparently located between the DH and ACP domains. The lack of KR domain suggests that the keto groups remain unreduced in the nascent polyketide chain. A possible mechanism for the formation of the aromatic ring has been suggested (Scheme 19). Four predicted enzymes that include a hydroxylase, a halogenase and two methyltransferases putatively modify the aromatic ring to furnish the final product. The predicted cytochrome P450 (CalO2) and FADH2-dependent halogenase (CalO3) are likely to catalyze the hydroxylation and iodination respectively, whereas the O-methyltransferases CalO1 and CalO6 install the two methyl groups. Although the precise timing of the post-PKS steps and the stringency of the substrate specificity of the enzymes remain to be fully established, biochemical and structural characterization of CalO2 indicated that the hydroxylation may occur after the halogenations. Based on the crystal structure of CalO2, MoCoy and coworkers also suggested that CalO2 may act on a CoA-derivatized or ACP domain-tethered substrate. Thorson and coworkers have also confirmed that sugar C is installed on the orsellenic group by glycosyltransferase CalG1. Meanwhile, the FabH/KS domainlike CalO4 is likely to catalyze the formation of the thioester linkage between the orsellenic acid and sugar B.
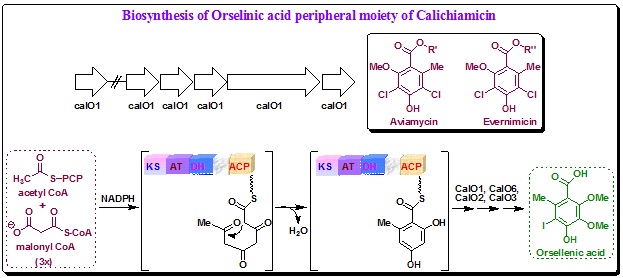 |
Scheme 19. Putative biosynthetic mechanism for the orsellenic acid moiety of calicheamicins. |