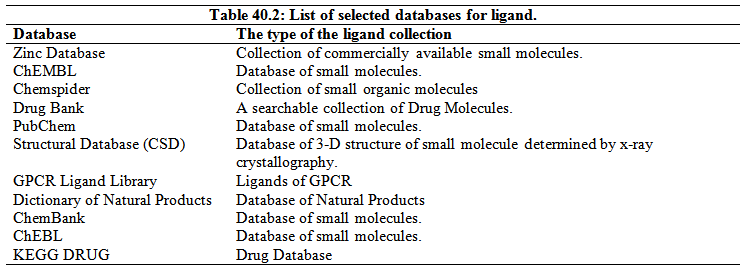
3. Collection of the inhibitor molecules- A list of selected database of ligand is given in Table 40.2. For most of these database, either keyword or the chemical structure can be used to search the database. The molecules from these database can be downloaded in the 2-D or 3-D conformation.
4. Docking-A list of molecular modeling and docking software are given in the Table 40.3.
Different steps in docking protocol: We will take the example of Autodock to understand different steps of docking. Autodock 4.1 is one of the most popular docking softwares. It has following steps to perform docking of a small molecules-
Step 1 and 2: Preparation of Macromolecule and Ligand for AutoDock- Step 1 and 2 are required to give the target and inhibitor molecule suitable environment for optimal docking. This step also allows to define the number of bonds can be made rotable for ligand to adopt suitable conformation for fitting within the binding pocket.
Step 3: Preparation of Grid Parameter file- This step allow to select the active site through drawing a grid of suitable size to define the space where a ligand molecule will be docked.
Step 4: Preparing the docking parameter files- This step allow to define the energy parameters and other docking parameters.
Step 5: Running of the docking