The Alignment of two query sequences can be global or local (Figure 39.4). In global alignment, the complete length of the protein sequences are compared to another where as in the case of local alignment, only a part of the sequence is compared (Figure 39.4). The global alignment is used to classify the protein into different classes where as local alignment is used to identify the motif or domain.
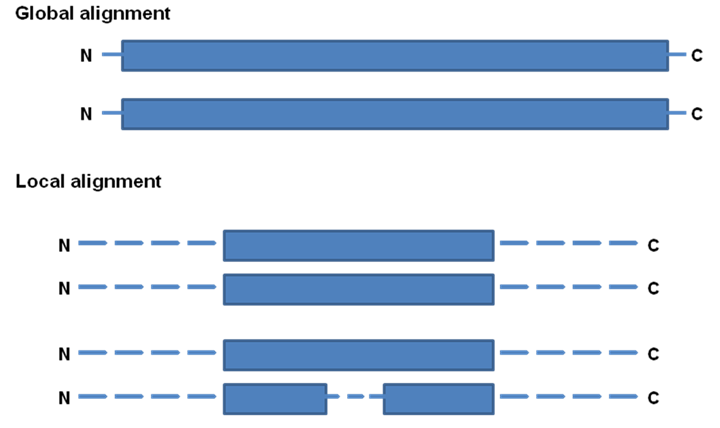
Figure 39.4: Sequence alignment of protein sequences.
To compare more than two sequences, multiple sequence alignment can be performed with ClustalW. It exploits the fact that similar sequences are usually homologous. First the pairwise alignment are carried out with the most similar sequences. Then based on the score of pairwise alignment, all sequences are classified into different groups. These groups are presented as multiple sequence alignment (Figure 39.5). As ClustalW calculates the distances between different sequences, it can be use to generate phylogenetic tree (Figure 39.6).