The use of a nucleotide scoring matrix to obtain optimal alignment of two nucleotide sequence is given in Figure 39.2. In this case, an identity matrix is relevant as the four nucleotide will not show any similarity to each other. As given the alignment examples, the sliding of the sequences gives different scores (3 or 7 using identity matrix and the alignment with the best score is choosen.
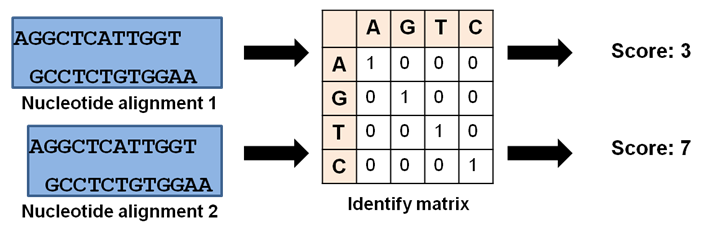
Figure 39.2: Sequence alignment of nucleotide sequences.
Opposite to the nucleotides, identity matrix is not sufficient to perform alignment of two protein sequences. Amino acids present in two sequences may have similar or different physiochemical properties. The probability to substitute one amino acid with other amino acids is also considered to give the score in the matrix (Figure 39.3). For example, aspartic acid is often observed with glutamic acid but substitution of aspartic acid with tryptophan is rare. This is due to the gentic codes of these amino acids ( aspartate and glutamic acid has only 3rd codon different) and their properties (both aspartate and glutamic are negatively charged amino acids). In addition, the effect of substitution on the protein structure is also been consider to provide score in the matrix. Asparate (negatively charged) to trptophan (aromatic) will have severe impact on the protein structure and hence will have lower score (In the matrix given in Figure 39.3, such a substitution will have -4 score). The most commonly used scoring matrix are the PAM (position assisted matrix) and BLOSUM (blocks substitution matrix). The negative value in the matrix indicate that the occurrence is coincidental where as positive values suggest a favorable substitution. In the example given in Figure 39.3, the two amino acid sequences are slide over to each other to produce two alignment. Using the blosum matrix, the amino acid alignment 1 is giving a