DNA polymerase I
- Not the primary enzyme for DNA synthesis: DNA polymerase I is so named because it was the first of the DNA polymerase enzymes to be isolated and characterized. Although it is capable of template-directed DNA synthesis, it is now known not to be the enzyme primarily responsible for new DNA synthesis, but it does have other important roles, as described below.
- 5' to 3' exonuclease activity: DNA polymerase I has an unusual 5' to 3' exonuclease activity. This gives it the ability to start at a single-stranded break and progressively remove nucleotides and replace them in a 5' to 3' direction.
- Removal of RNA primer: In the lagging strand, when DNA polymerase III runs into the 5'-end of the previous RNA primer, it is unable to proceed further. Although the new DNA butts up against the primer, it is not covalently joined to it. DNA polymerase III dissociates, leaving a "nick" (a single stranded gap) between the new DNA and the primer. Starting at the nick, DNA polymerase I removes the primer ribonucleotides one at a time, using its 5' to 3' exonuclease activity, and replaces them with deoxyribonucleotides, using its DNA polymerase activity.
- DNA repair: DNA polymerase I is also used to fill in short gaps in DNA, often as a part of a repair process that excises part of a damaged strand and replaces it with new DNA templated from the remaining strand.
Proofreading and DNA repair
- Removal of mismatched bases: DNA polymerases III and I both have 3' to 5' exonuclease activity. This allows them to remove a mismatched nucleotide that has just been added to a growing DNA chain and make another attempt to insert the correct nucleotide. This "proofreading" function helps to reduce the number of mistakes in DNA synthesis that would otherwise result in mutation.
- DNA polymerase II: There is yet another prokaryotic DNA polymerase, designated DNA polymerase II, whose main function appears to be to synthesize replacement DNA during DNA repair.
Final steps in DNA synthesis
- DNA Ligase: After the last ribonucleotide is removed from the primer and replaced with a deoxynucleotide, there is still a nick in the newly synthesized lagging strand. This nick is closed by DNA ligase, which forms a covalent phosphodiester bond between the Okazaki fragments, joining them into a continuous strand of DNA. Note that Okazaki fragments accumulate when ligase function is impaired.
- Topoisomerase II: Its role is to cut and reseal the newly synthesized DNAs as needed so that they can separate from each other (it is easy to visualize two circular genomes linked through each other at the end of replication).
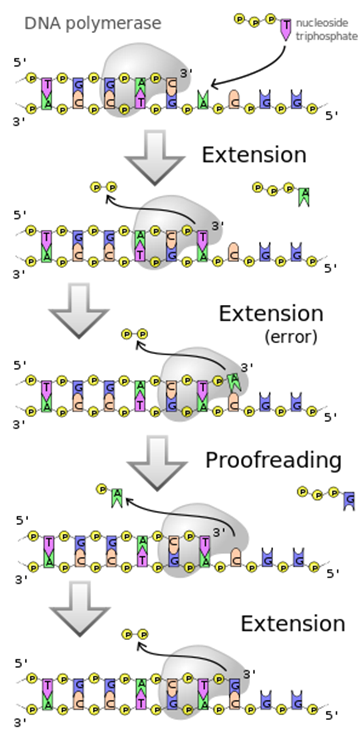
Fig. 7 . Diagram of DNA polymerase extending a DNA strand and proof-reading.
REFERENCES:
Text Books:
1. Jeffery C. Pommerville. Alcamo's Fundamentals of Microbiology (Tenth Edition). Jones and Bartlett Student edition.
2. Gerard J. Tortora, Berdell R. Funke, Christine L. Case. Pearson - Microbiology: An Introduction. Benjamin Cummings.
3. J. Krebs, E.S. Goldstein, Stephen T. Kilpatrick. Lewin's Genes X. Jones and Bartlett Publishers.
Reference Books:
1. Lansing M. Prescott, John P. Harley and Donald A. Klein. Microbiology. Mc Graw Hill companies.