DNA replication
Templated replication
- Sense and antisense strands: The sense strand of a double-stranded DNA molecule has a base sequence similar to that of the RNA that is transcribed from the DNA. The antisense strand (also known as template strand) has a sequence that is the reverse complement of the sense strand (base paired to it with an antiparallel 5' to 3' polarity).
- Semiconservative replication: Discovery of the double helical structure of DNA immediately suggested a mechanism for precise replication -- namely separation of the two strands and template synthesis of a new copy of each of the missing complementary strands. This model predicted that each new double helix should have one strand of parental DNA and one strand of newly synthesized DNA.
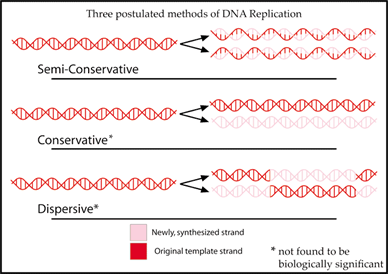
Fig 6. Three postulated methods of replication: 1. Semiconservative, 2. Conservative, 3. Dispersive
- Messelson-Stahl experiment: Semiconservative replication was demonstrated by starting with bacteria whose DNA was labeled with heavy nitrogen (15 N) and then allowing various amounts of growth to occur in media labeled with normal nitrogen. After one round of replication, the entire DNA had an intermediate density. After two rounds, one half was light and the other half still had an intermediate density. There was no conservation of heavy DNA. However, the data suggested that each of the labeled strands from the original DNA remained intact and separated from its partner in each replication cycle.
Initiation of replication
- Origin of Replication: The DNA of a bacterial chromosome is a closed circular structure. DNA replication begins at a specific site (origin of replication) characterized by the presence of repeated 9 base and 13 base nucleotide sequences. The repeating units are referred to as 9-mers and 13-mers. The 13 mers are AT rich, making easier to separate the two strands of the double-stranded DNA.
- DNAa protein: The protein coded by the DNA a gene binds to the repeated 9mers. This forms a tight loop and generates a strain that causes strand separation in the region containing the AT-rich 13-mers
- Helicase: Enzymes called helicases use energy derived from ATP to further separate the two strands of the DNA double helix.
- Topoisomerase I: Separation of the two strands of the DNA double helix requires substantial unwinding of the helix. An enzyme known as topoisomerase I (gyrase) relieves twisting strain that is generated by unwinding the double helix. It is believed to act by cutting one of the strands such that the other strand can rotate freely to relieve the strain, and then resealing the strand that has been temporarily cut.
- Keeping the helix open: Single stranded DNA-binding proteins (SSBPs) attach to the single stranded DNA generated by unwinding the double helix and temporarily keep it from reforming double helical structures.
Patterns of replication
- Replication forks: When the two strands of double helical DNA separate and replication of both strands begins, a forked or Y-shaped structure is formed.
- Bidirectional replication: In bacterial cells, replication starts at a specific origin of replication within the circular DNA molecule and proceeds in both directions away from the origin. This results in the formation of a replication "bubble", which continues to elongate as replication proceeds. A similar pattern is seen in eukaryotic chromosomes, except that multiple origins are involved.
- Theta structure in bacteria: As replication proceeds in both directions around the circular chromosomes of bacteria, a structure reminiscent of the Greek letter theta is formed.
- Replicons: The very long DNA double helices found in eukaryotic chromosomes contain multiple origins of replication, which often initiate bidirectional replication more or less synchronously. Each unit of replication is called a replicon. Replication continues until the replication bubbles fuse to yield fully replicated DNA strands.
- Other patterns There are also two alternative strategies for replication of circular DNA that are more complex than the simple bidirectional model. These are the rolling circle mode (sigma mode) used by some types of viruses, and the D loop mode, used for replication of mitochondrial and chloroplast DNAs.
5'-to-3' synthesis
- Unidirectional addition of nucleotides: Polymerization of DNA, and also of RNA, occurs by condensing a 5'-nucleoside triphosphate (dNTP or NTP) onto the 3' hydroxyl group of another nucleotide, or onto the 3'-end of a growing polynucleotide chain. No mechanism exists for extending the 5'-end of a nucleotide chain.
- Energy for synthesis: Hydrolysis of the dNTP or NTP that is being added provides the energy needed to form a covalent bond. The outer two phosphates of the triphosphate are split off and the innermost phosphate forms an ester linkage with the 3'-hydroxyl group at the end of the pre-existing chain. This results in a phosphodiester bond