Maturation process involves methylation of 30s rRNA precursor at specific sites occurring at 2’hydroxyl groups of bases. Some bases such as uridine is modified to pseudouridine or dihydrouridine. Further cleavage process is carried out using enzymes RNase III, RNase P, and RNase E at sites 1, 2 and 3 respectively as shown in the Figure 29.5. Intermediate products are formed namely 17s, tRNA, 25S and 5S. These are acted on by certain nucleases to give final products of 16s, tRNA, 23s, 5s rRNA respectively which is given in figure 29.5.
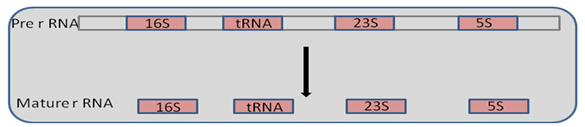
Figure 29.5:RNA splicing in Bacteria
rRNA processing in Vertebrates (Eukaryotes): In Eukaryotes nucleolus is the centre of processing ribosomal RNA. A 45s precursor is formed by RNA polymerase I and processed in 90s preribosomal nucleolar complex to give 18s, 28s, and 5.8s rRNA which described in Figure 29.6. There is tight coupling of RNA processing with ribosomal assesmbly.5s rRNA is transcribed by RNA polymerase III from a separate gene. Precursor RNA undergoes methylation at more than 100 bases from 14000 nucleotides at 2’ hydroxyl group. Furthermore there is modification of bases such as uridine to pseudouridine etc. followed by series of cleavage reaction. Cleavage and modifications are guided by snoRNAs (small nucleolar RNA). In yeast, entire processing involves pre-RNA, 170 non-ribosomal protein, 70 snoRNA and 78 ribosomal proteins. snoRNA are supposed to be remnant of spliceosomes.
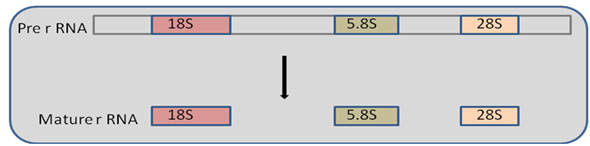
Figure 29.6:RNA splicing in Eukaryotes