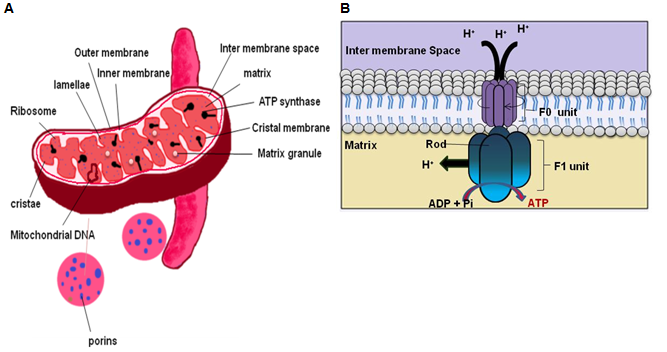
Figure 22.3: Mitochondria. (A) Struture of mitochondria and (B) enlarged view of ATP Synthase.
Porin allows free movement of molecules less than 5000da within and outside mitochondria. Where as larger molecules or proteins moves into the mitochondria through trasnporters involving signal peptides known as “mitochondrial targeting sequence”. Inner membrane is folded into membrane projections to form cristae. Cristae occupies major area of membrane surface and house machinary for anaerobic oxidation and electron transport chain to produce ATP. Due to presence of inner and outer membrane, mitochondria can be divided into 2 compartments: first in between the inner and outer membrane, known as intermembrane space and second inside the inner membrane known as matrix. The proteins present in intermembrane space have a role in executing “programmed cell death” or “apoptosis”. Matrix is the liquid part present in the inner most of mitochondria and it contains ribosome, DNA, RNA, enzymes to run kreb cycle and other proteins. Mitochondrial DNA is circular and it has full machinery to synthesize its own RNA (mRNA, rRNA and t-RNA) and proteins. A number of difference exist between mitochondrial DNA and DNA present in nucleus and these difference are not discussed here due to space constrain. Electron transport chain components (complex I to complex V) are integral proteins, present in the inner membrane of mitochondria. During metabolic reactions such as glycolysis, kreb cycle [metabolic reaction are discussed later] produces large amount of reducing equivalent in the form of NADH2 and FADH2. Electron transport chain process reducing equivalent and flow of the electron through different complexes (Complex I to Complex IV) causes generation of proton gradient across the membrane.