2.4.4. Biosynthesis of C-1027
The C-1027 chromophore can be dissected into 4 biosynthetic building blocks which are
(a) an enediyne core - synthesized via polyketide pathway by enediyne PKSs.
(b) a β-amino acid - annotation of the gene products of the C-1027 gene cluster suggested that the β-amino acid is derived from L-tyrosine.
(c) a deoxy aminosugar - initial annotation of the gene products of the C-1027 gene cluster suggested that the deoxy amino sugar is derived glucose-1-phosphate.
(d) a benzoxazolinate moiety -The biosynthesis of the benzoxazolinate moiety was originally proposed to start from anthranilate, a common intermediate from the shikimate pathway. Subsequent characterization of the enzymes from the putative pathway revealed an unexpected pathway with chorismic acid as the starting material (Figure 14)
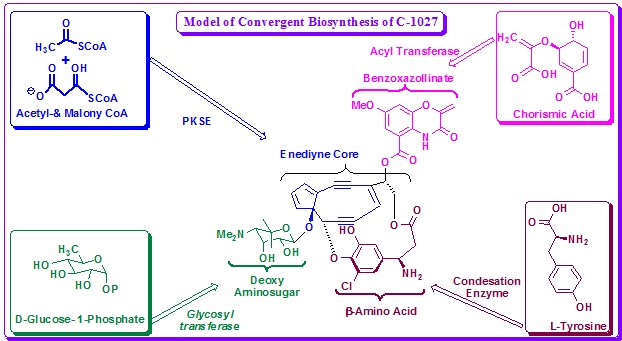
Sequencing of the gene cluster revealed homologs for a glycosyl transferase, an acyltransferase, and condensation enzymes, suggesting a convergent biosynthetic approach is used in enediyne assembly for C-1027. The initial enzymatic steps in every biosynthetic pathway for C-1027 have been analyzed using a combination of in vivo gene inactivation and in vitro characterization of recombinant enzymes. This experiment established the starting metabolite for each moiety and provided substantial evidence for a convergent approach in enediyne biosynthesis.
Enediyne core (4) biosynthesis: There are 13 genes identified within the C-1027 gene cluster that are consistent with the structure of C-1027 and are essential for the production of enediyne core of C-1027. These are sgcE, sgcE1, sgcE2, sgcE3, sgcE4, sgcE5, sgcE6, sgcE7, sgcE8, sgcE9, sgcE10, sgcE11 and sgcF that encode the enediyne core (4) biosynthesis (Figure 15).
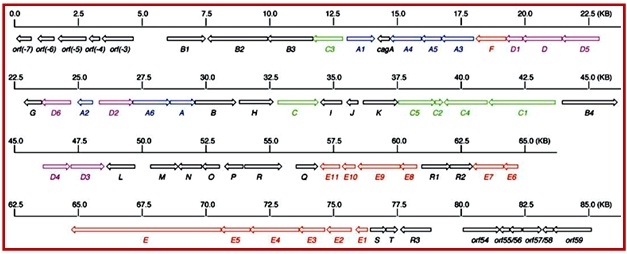 |
Figure 15. Organization of the C-1027 biosynthesis gene cluster ORFs outside the sgcB1 to sgcR3 region are not essential for C-1027 production. Color coding indicates the biosynthesis genes for the enediyne core (red), deoxyaminosugar (blue), β-amino acid (green), benzoxazolinate (purple), and all other genes (black). (Liu, W.; Christenson, S. D.; Standage, S.; Shen, B.Science 2002, 297, 1170.) |
It is not clear whether the enediyne cores are assembled by de novo polyketide biosynthesis or degradation from a fatty acid precursor. However, feeding experiments with 13C-labeled precursors supported that both the nine- and 10-membered enediyne cores are derived from a minimum of eight acetate units organized in head-to-tail. Importantly, only one gene, sgcE, among all other genes indentified within the C-1027 cluster encodes a PKS. SgcE contains five domains—(a) the ketoacyl synthase (KS), (b) acyltransferase (AT), (c) ketoreductase (KR), (d) dehydratase domains that are characteristic of known PKSs, and (e) a domain at the COOH-terminus that is unique to enediyne PKSs. Moreover a putative acyl carrier protein domain is proposed to present in between AT and KR region (Figure 16). Most importantly, the SgcE enediyne PKS exhibits head-to-tail sequence homology. It has an identical domain organization to the CalE8 enediyene PKS that catalyzes the biosynthesis of the 10-membered endiyne core of calicheamicin in Micromonosporaechinospora. Therefore, the nine- and 10-membered enediyne cores share a common polyketide biosynthetic pathway.
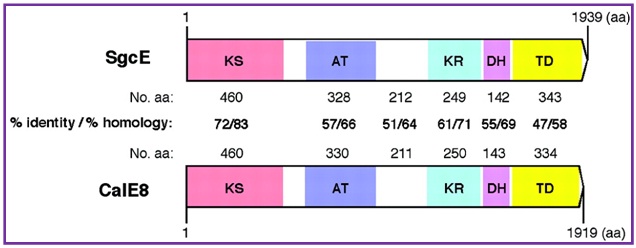 |
Figure 16. Comparison between the SgcE PKS catalyzing the nine-membered enediyne core in C-1027 biosynthesis and the CalD8 PKS catalyzing the 10-membered enediyne core in calicheamicin biosynthesis. aa = amino acid; KS = ketoacyl synthase; AT = acyltransferase; ACP = acyl carrier protein; KR = ketoreductase; DH = dehydratase; and TD = COOH-terminal domain. (Liu, W.; Christenson, S. D.; Standage, S.; Shen, B. Science 2002, 297, 1170.) |
It is proposed that the SgcE catalyzes the assembly of a nascent linear polyunsaturated intermediate from acetyl and malonyl coenzyme A (CoA) in an iterative process. Next, upon action of other enzymes polyunsaturated intermediate is subsequently desaturated to furnish the two -yne groups and cyclized to afford the enediyne core. An acetylenase has been reported from the plant Crepisalpina that was characterized as a nonheme di-iron protein. However, no such homolog was found within the C-1027 cluster. Close comparison of the C-1027 gene cluster with that of neocarzinostatin gene cluster revealed the presence of a group of ORFs (sgcE1 to sgcE11) in addition to sgcE. These ORFs are highly conserved.
The functions of the genes are as follows: (a) Genes SgcE6, SgcE7 and SgcE9 function similar to that of various oxidoreductases. (b) SgcE1, SgcE2, SgcE3, SgcE4, SgcE5, SgcE8 and SgcE11 show either no sequence homology or homology only to proteins of unknown function. (c) SgcE10 is highly homologous to a family of thioesterases. These enzymes, together with the SgcF epoxide hydrolase are responsible for the synthesis of enediyne intermediate 4 from the nascent linear polyunsaturated intermediate (Scheme 7). All other experimental evidences support the polyketide route to the biosynthesis of C-1027 enediyne core.
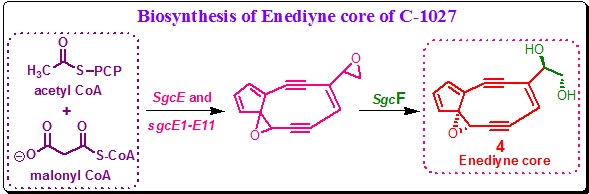 |
Scheme 7. Biosynthetic hypothesis for the enediyne core and a convergent assembly strategy. |
Deoxyaminosugar (5) biosynthesis: There are seven genes namely, sgcA, sgcA1, sgcA2, sgcA3, sgcA4, sgcA5, sgcA6 that are responsible for the biosynthesis deoxyaminosugar (5) moiety in C-1027. The seven deoxyaminosugar biosynthesis genes encode a thymine diphosphate (TDP)-glucose synthetase (SgcA1), a TDP-glucose 4,6-dehydratase (SgcA), a TDP-4-keto-6-deoxyglucose epimerase (SgcA2), a C-methyl transferase (SgcA3), an amino transferase (SgcA4), an N-methyl transferase (SgcA5) and a glycosyltransferase (SgcA6). Together, they do the enzyme functions that is essential for the biosynthesis of 5 from glucose-1-phosphate (Scheme 8) and the attachment of 5 to 4.
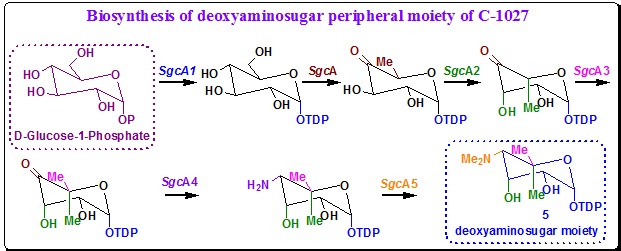 |
Scheme 8. Biosynthetic hypothesis for deoxyaminosugar (5, C). |
β-amino acid (6) biosynthesis: There are six genes, sgcC, sgcC1, sgcC2, sgcC3, sgcC4, sgcC5 thatencode biosynthesis of β-amino acid (6) in C-1027. The six β-amino acid biosynthesis genes encode (a) a phenol hydroxylase (SgcC), (b) a non ribosomal peptide synthetase (NRPS)–adenylation enzyme (SgcC1), (c) an NRPS peptidyl–carrier protein (PCP) (SgcC2), (d) a halogenase (SgcC3), (e) an aminomutase (SgcC4), and (f) an NRPS-condensation enzyme (SgcC5). These enzyme together functions for the biosynthesis of β-amino acid (6) moiety starting from tyrosine (Scheme 9). Once formed it is activated by SgcC5 as an aminoacyl-S-PCP. The activated aminoacyl-S-PCP is then ready for attachment to the enediyne moiety 4 by SgcC5.
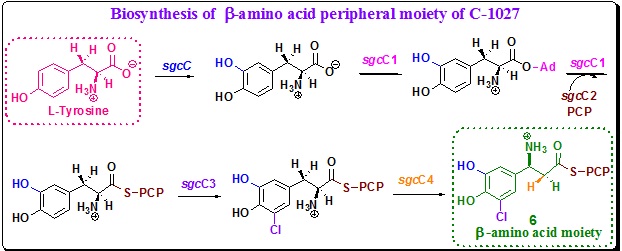 |
Scheme 9. Biosynthetic hypothesis for β-amino acid (6, D). |
Benzoxazolinate (7) biosynthesis: Seven genes were identified that are essential for the biosynthesis of benzoxazolinate moiety of C-1027. These are sgcD, sgcD1, sgcD2, sgcD3, sgcD4, sgcD5, sgcD6. These seven benzoxazolinate biosynthesis genes encode the followings: (a) the anthranilate synthase I and II subunits (SgcD and SgcD1), (b) a monoxygenases (SgcD2), (c) a P-450 hydroxylase (SgcD3), (d) an O-methyl transferase (SgcD4), (e) a CoA ligase (SgcD5), and (f) an acyltransferase (SgcD6). Action of these enzymes starts with anthranilate, a common intermediate from the shikimate pathway, for the biosynthesis of benzoxazolinate moiety (7) (Scheme 10). The co-localization of SgcD and SgcD1 gene along with the other C-1027 production genes assures the availability of anthranilate for secondary metabolite biosynthesis. However, the origin of the C3 unit is not clear. Moreover, how the C3 unit is fused to the anthranilate intermediate to form the morpholinone unit of benzoxazolinate moiety (7) is also unclear. However, it is believed that the anthranilate unit of benzoxazolinate moiety (7) is activated as an acyl-S-CoA for attachment to the enediyne core 4 by the action of gene SgcD6 (Scheme 11).
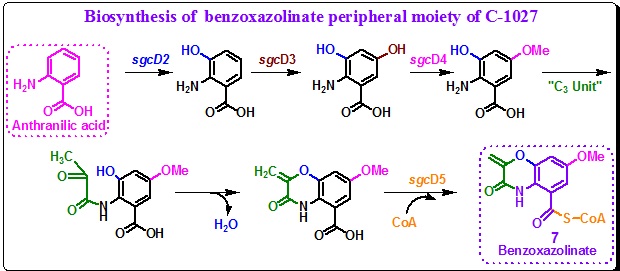 |
Scheme 10. Biosynthetic hypothesis for benzoxazolinate (7, E). |
Biosynthesis of C-1027 by attaching the peripheral moieties to the enediyne core:
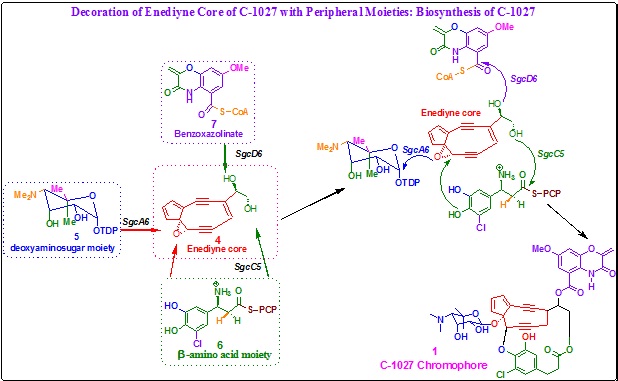 |
Scheme 11. Final step in biosynthesis of C-1027-decoration of enediyne core of C-1027 with peripheral moieties. |