2.4.3.2 General Biosynthesis of Enediyne Cores
Though there exists various types of gene clusters for different enediynes, a unified biosynthetic scheme for the nine- and ten-membered enediyne cores is now possible owing to the discovery of a shared iterative type I PKS (polyketide synthase). Bacterial polyketide biosynthesis carries out either by noniterative, modular type I PKS, or a multienzyme complex of iteratively acting type II PKS, or homodimeric, iteratively acting condensing type III PKS without an acyl carrier protein (ACP).
In contrast, the enediyne PKSs from the biosynthetic pathways of calicheamicin ![]() and C-1027 seem to resemble a family of fungal iterative type I PKSs. To date, five gene clusters cloned only one type of PKS that are all iterative type I PKS. The gene cluster for C-1027 contains a single PKS. This PKS is shared by sequence homology and domain architecture among the enediyne family (Fig. 4A). Disruption of this SgcE (the PKS for C-1027 synthesis) in the producer of C-1027 did not produce C-1027. Identical results were observed with the homologous neocarzinostatin PKS, NcsE. These results provide strong support that the enediyne core is produced by a polyketide pathway. The enediyne PKSs use a single set of catalytic domains for polyketide synthesis. Bioinformatic analysis of this PKS family, PKSE, showed that the enediyne PKS encompass four domains in the following order (N- to C-terminus): (a) a ketosynthase (KS), (b) an acyltransferase (AT), (c) a ketoreductase (KS), and (d) dehydrogenase (DH). Enediyne PKS possess closest sequence homology to polyunsaturated fatty acid (PUFA) synthases involved in the biosynthesis of docosahexaenoic acid in Moritella marina and eicosapentaenoic acid in Shewanella (Figure 10). The significant sequence identity and similarity shared by PKSs, CalE8 (the PKS for calicheamicin synthesis) and SgcE (the PKS for C-1027 synthesis) indicated that the 9- and 10-membered enediyne cores may share a common PKS progenitor. Thus, the iterative type I PKS is unique to all of the enediyne family. It is now evident that the enediyne biosynthetic gene clusters share conserved architecture featuring the enediyne PKS. These enediyne PKS formed the basis of several expedient strategies to clone additional enediyne biosynthetic gene clusters such as of NCS, maduropeptin, and dynemicin gene clusters via a PCR approach.
and C-1027 seem to resemble a family of fungal iterative type I PKSs. To date, five gene clusters cloned only one type of PKS that are all iterative type I PKS. The gene cluster for C-1027 contains a single PKS. This PKS is shared by sequence homology and domain architecture among the enediyne family (Fig. 4A). Disruption of this SgcE (the PKS for C-1027 synthesis) in the producer of C-1027 did not produce C-1027. Identical results were observed with the homologous neocarzinostatin PKS, NcsE. These results provide strong support that the enediyne core is produced by a polyketide pathway. The enediyne PKSs use a single set of catalytic domains for polyketide synthesis. Bioinformatic analysis of this PKS family, PKSE, showed that the enediyne PKS encompass four domains in the following order (N- to C-terminus): (a) a ketosynthase (KS), (b) an acyltransferase (AT), (c) a ketoreductase (KS), and (d) dehydrogenase (DH). Enediyne PKS possess closest sequence homology to polyunsaturated fatty acid (PUFA) synthases involved in the biosynthesis of docosahexaenoic acid in Moritella marina and eicosapentaenoic acid in Shewanella (Figure 10). The significant sequence identity and similarity shared by PKSs, CalE8 (the PKS for calicheamicin synthesis) and SgcE (the PKS for C-1027 synthesis) indicated that the 9- and 10-membered enediyne cores may share a common PKS progenitor. Thus, the iterative type I PKS is unique to all of the enediyne family. It is now evident that the enediyne biosynthetic gene clusters share conserved architecture featuring the enediyne PKS. These enediyne PKS formed the basis of several expedient strategies to clone additional enediyne biosynthetic gene clusters such as of NCS, maduropeptin, and dynemicin gene clusters via a PCR approach.
All the studies towards polyketide biosynthesis by enediyne PKSs established that with self-phosphopanetheinylation of a unique ACP, PKSE catalyses the enediyne synthesis. Therefore, enediyne biosynthesis follows an ACP-dependent PKSE-catalyzed pathway (Figure 11).
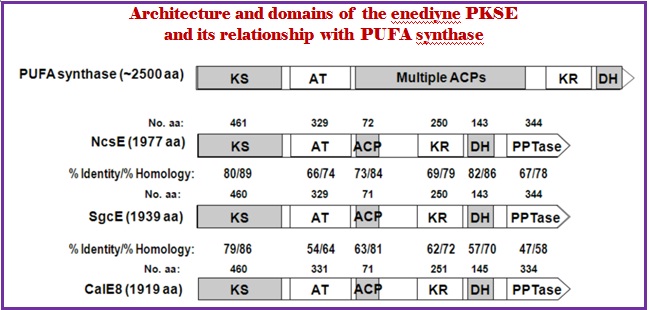 |
Figure 10. Architecture and domains of the enediyne PKSE and its relationship with PUFA synthase (Lanen, S. G. V.; Shen, B. Curr Top Med Chem. 2008, 8, 448–459.).
|
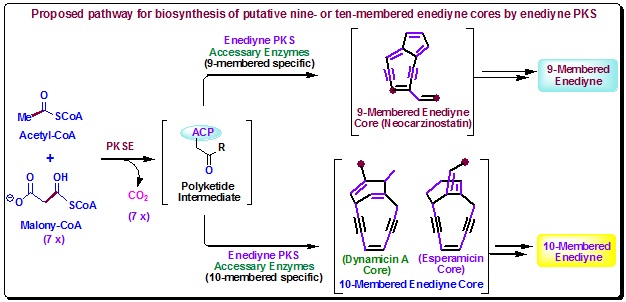 |
Figure 11. Proposed pathway for biosynthesis of a polyunsaturated intermediate from acyl CoA by PKSE and the subsequent transformation by enediyne PKS associated enzymes into putative nine- or ten-membered enediyne cores that are finally tailored to individual enediyne natural product. Atoms that were incorporated intact from acyl CoA precursors to the enediyne cores are shown in bold. |