4.20.7. Microarray Based DNA Detection
4.20.7.1. Introduction
A DNA microarray also known as gene chips, DNA chip, or biochip is a collection of microscopic DNA spots attached to a solid surface. DNA microarrays are used to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as probes or reporters. A probe can be a short section of a gene or other short DNA sequence that are used to hybridize a complementary DNA (cDNA) or cRNA sample that are called target under high-stringency conditions. The Probe-Target Hybridization event is usually detected by a quantifiable signal generation from the fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative nucleic acid sequences in the target (Figure 4.50). Since an array can contain tens of thousands of probes, a microarray experiment can accomplish many genetic tests in parallel. Therefore arrays have dramatically accelerated many types of investigation in genomics.
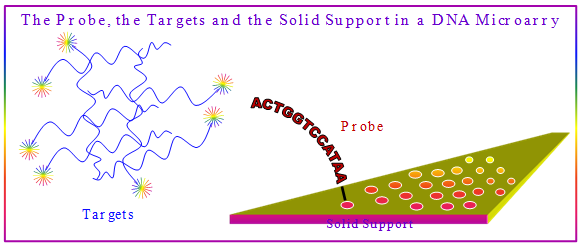
Figure 4.50: The definition of Target and Probe in a Microarry.
In standard microarrays, the probes are synthesized and then attached to a solid surface by a covalent bond to a chemical matrix such as epoxy-silane, amino-silane, lysine, polyacrylamide or others. The solid surface in general is a glass or a silicon chip, in which case they are collectively known as an Affy chip when an Affymetrix chip is used. Other microarray platforms, such as Illumina, use microscopic beads, instead of the large solid support. Alternatively, microarrays can be constructed by the direct synthesis of oligonucleotide probes on solid surfaces. DNA arrays are different from other types of microarray only in that they either measure DNA or use DNA as part of its detection system.
4.20.7.2. Uses of Microarray in DNA Detection
- DNA microarrays can be used to measure changes in expression levels, to detect single nucleotide polymorphisms (SNPs), or to genotype or resequence mutant genomes.
- Revolution in the analysis of genetic information
- Hybridization is a highly parallel search by each molecule for matching partner on an affinity matrix.
- Specificity and affinity of complementary base pairing.
- Use of glass as a substrate, fluorescence for detection and the development of new technologies for synthesizing or depositing DNA have allowed the miniaturization of DNA arrays with increases in information content.
- "DNA chip”/ "DNA microarray" allows to collect more information about DNA sequences in an afternoon than an army of scientists could collect in years using earlier several techniques.
- DNA chips promise to carry the science of understanding genomes to a whole new level, and to bring tools for getting DNA-sequence information out of research labs into doctors' offices, the better to tailor-fit medical treatments to an individual's particular genetic makeup.
- Principle Major technologies
- cDNA probes (> 200 nt), usually produced by PCR, attached to either nylon or glass supports
- Oligonucleotides (25-80 nt) attached to glass support
- Oligonucleotides (25-30 nt) synthesized in situ on silica wafers (Affymetrix)
- Probes attached to tagged beads.
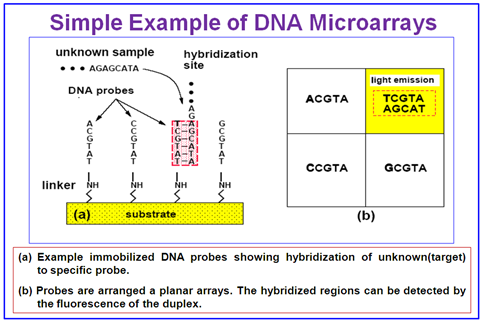
Figure 4.51: A simple example of DNA Microarray.