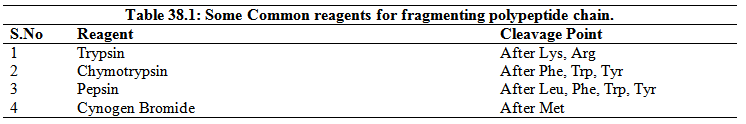
Stage 2. Cleavage of the polypeptide chain: Proteases and the chemical agents targeting proteins have a specific recognition sequence and they cleave after a particular amino acid. A list of protease and chemicals commonly used to digest the polypeptides into the small peptide fragment is given in Table 38.1.
Stage 3. Sequencing the peptides-Once the peptide fragments are generated, we can start the sequencing of each polypeptide chain. It has following steps:
A. Identifying the N-terminal residue: The N-terminal amino acid analysis is a 3 steps process.
1. Derivatization of terminal amino acid-The chemical reaction is performed to labeled terminal amino group with compounds such as sanger reagent 1-fluoro-2,4-dinitrobenzene (DFNB) and dansyl chloride. In most of the case these reagents also label free amino group present on basic amino acids such as lysine and arginine. In a reaction mechanism given in Figure 38.3, dinitrofluorobenzene reacts with the free amine group to form dinitrophenyl-amino acid complex.
2. hydrolyse the protein-Acid hydrolysis of dinitrophenyl-amino acid complex leads to the breaking of peptide bond to release dinitrophenyl-amino acid complex in solution.
3. Separation and analysis of derivatized amino acids-A HPLC or TLC separation of complex and comparing with the standard amino acids.
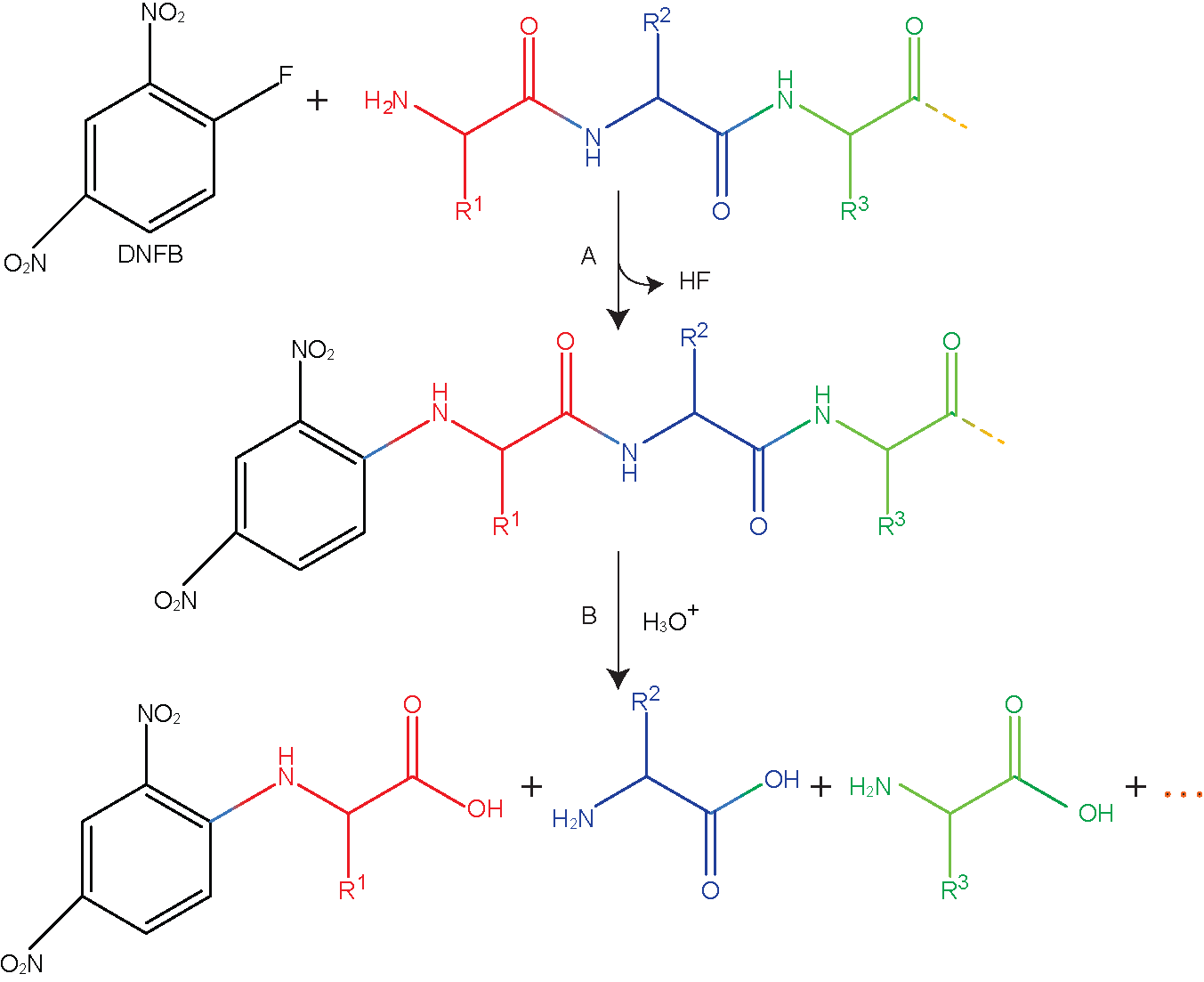
Figure 38.3: Derivatization of N-terminal amino acid with sanger reagent.