3-6.2.11 Transposon mediated mutagenesis
Transposons are DNA elements which have ability to move from one region to another region in the genome. The shifting of transposon element from a heterochromatin region to a transcriptionally active region results in insertional mutagenesis. It can be exploited as a genetic tool for analysis of gene and protein function. The study of transposons is well described in the case of Drosophila (in which P elements are most commonly used) and in Thale cress ( Arabidopsis thaliana ) and bacteria such as Escherichia coli .
Transposon based mutagenesis is a biological process that helps in genes to be transferred to a host organism's chromosome, modifying the function of an extant gene on the chromosome and causing mutation. Transposon mutagenesis was first studied by Barbara McClintock in the mid-20th century with corn for which she was awarded Nobel prize also.
In case of bacteria, transposition mutagenesis is usually done by the help of a plasmid from which a transposon is extracted and inserted into the host chromosome. This usually requires a set of enzymes including transposase to be translated.
Currently transposons can be used in genetic research and recombinant genetic engineering for insertional mutagenesis. Insertional mutagenesis is when transposons act like vectors to help remove and integrate genetic sequences. Due to their simple and intricate design and inherent ability to move DNA sequences, transposons are highly compatible for transducing genetic material, enable them as an ideal genetic tool .
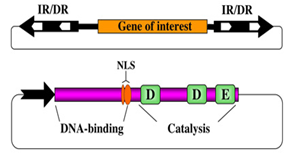
Fig 3-6.2.11 [A]
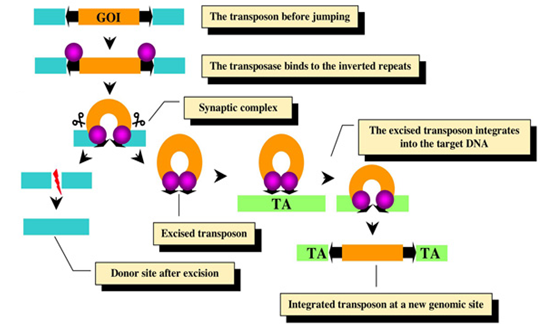
Fig 3-6.2.11 [B]
Fig 3-6.2.11: The Sleeping Beauty transposon system.
(Source: https://www.mdc-berlin.de/8201913/en/research/research_teams/mobile_dna/research)
Figure [A] describes the components and structure of a two-component gene transfer system based on Sleeping Beauty. A gene of interest (orange box) to be mobilized is cloned between the terminal inverted repeats (IR/DR, black arrows) that contain binding sites for the transposase (white arrows). The transposase gene (purple box) is physically separated from the IR/DRs, and is expressed in cells from a suitable promoter (black arrow). The transposase consists of an N-terminal DNA-binding domain, a nuclear localization signal (NLS) and a catalytic domain characterized by the DDE signature.
Fig [B] illustrates the mechanism of Sleeping Beauty transposition system. The transposable element carrying a gene of interest (GOI, orange box) is maintained and delivered as part of a DNA vector (blue DNA). The transposase (purple circle) binds to its sites within the transposon inverted repeats (black arrows). Excision takes place in a synaptic complex. Excision separates the transposon from the donor DNA, and the double-strand DNA breaks that are generated during this process are repaired by host factors. The excised element integrates into a TA site in the target DNA (green DNA) that will be duplicated and will be flanking the newly integrated transposon.