-2.3.2.3 Annealing:
Following the separation of the two strands of DNA during denaturation, the temperature of the reaction mix is lowered to 50–65°C for 20–50 seconds to allow annealing of the primers to the single-stranded DNA templates. Typically the annealing temperature should be about 3-5°C below the Tm of the primers. Stable complimentary binding are only formed between the primer sequence and the template when there is a high sequence complimentarity between them. The polymerase enzymes initiate the replication from 3' end of the primer towards the 5'end of it.
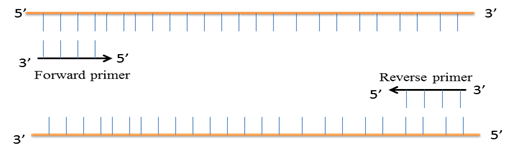
Fig 3-2.3.2.3 : Annealing of primer
3-2.3.2.4 E xtension /e longation:
Extension/elongation step includes addition of dNTPs to the 3' end of primer with the help of DNA polymerase enzyme. The type of DNA polymerase applied in the reaction determines the optimum extension temperature at this step. DNA polymerase synthesizes a new DNA strand complementary to its template strand by addition of dNTPs, condensing the 5'-phosphate group of the dNTPs with the 3'-hydroxyl group at the end of the nascent (extending) DNA strand. Conventionally, at its optimum temperature, DNA polymerase can add up to a thousand bases per minute. The amount of DNA target is exponentially amplified under the optimum condition of elongation step. The drawback of Taq polymerase is its relatively low replication fidelity. It lacks a 3' to 5' exonuclease proofreading activity, and has an error rate measured at about 1 in 9,000 nucleotides.
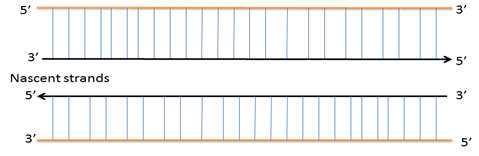
Fig 3-2.3.2.4 : Extension/ Elongation