1-3.7.1 Examples of Cloning Vector:
1-3.7.1.a pBR322
• pBR322 is a widely-used E. coli cloning vector. It was created in 1977 in the laboratory of Herbert Boyer at the University of California San Francisco. The p stands for " plasmid " and BR for "Bolivar " and " Rodriguez ", researchers who constructed it.
• pBR322 is 4361 base pairs in length.
• pBR322 plasmid has the following elements:
► “ rep ” replicon from plasmid pMB1 which is responsible for replication of the plasmid.
► “ rop ” gene encoding Rop protein. Rop proteins are associated with stability of RNAI-RNAII complex and also decrease copy number. The source of “ rop ” gene is pMB1plasmid.
► “ tet ” gene encoding tetracycline resistance derived from pSC101 plasmid.
► “ bla ” gene encoding β lactamase which provide ampicillin resistance (source: transposon Tn3).
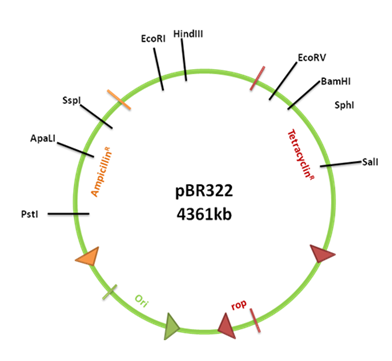
Fig 1-3.7.1.A: Plasmid pBR322.
1-3.7.1.b pUC plasmids:
• pUC plasmids are small, high copy number plasmids of size 2686bp.
• This series of cloning vectors were developed by Messing and co-workers in the University of California. The p in its name stands for plasmid and UC represents the University of California.
• pUC vectors contain a lacZ sequence and multiple cloning site (MCS) within lacZ. This helps in use of broad spectrum of restriction endonucleases and permits rapid visual detection of an insert.
• pUC18 and pUC19 vectors are identical apart from the fact that the MCS is arranged in opposite orientation.
• pUC vectors consists of following elements:
► pMB1 “rep ” replicon region derived from plasmid pBR322 with single point mutation (to increase copy number).
► “ bla ” gene encoding β lactamase which provide ampicillin resistance which is derived from pBR322. This site is different from pBR322 by two point mutations.
► E.coli lac operon system.
• “rop ” gene is removed from this vector which leads to an increase in copy number.