Recognition sequences for type II restriction endonucleases:
Type II restriction endonucleases are homodimeric polypeptide. These homodimer enzymes recognize short nucleotide sequences of about 4-8 bp known as restriction site and are usually palindromic in nature (Fig. 2). Most of the restriction enzymes used in molecular biology research are six base cutters. Restriction enzymes such as Eco R1, cuts the double stranded DNA at its recognition site in which the single stranded complementary tails called "sticky" or cohesive ends are generated. These single stranded sticky ends can form hydrogen bond with the complementary DNA sequence from different source. For example, two DNA sequences of different origin both containing Eco R1 restriction site can be ligated if they are digested with the Eco R1 restriction enzyme, as both produce sticky ends that are complementary to each other (Fig. 3). Some of the type II restriction endonucleases, such as sma I, cuts double strand DNA at the same position and generate blunt ends when they cleave the DNA. Restriction endonucleases show high degree of specificity in recognizing specific restriction site which is specific for that particular restriction enzyme. Any change in recognition site of the restriction endonuclease essentially eliminates total enzymatic activity.
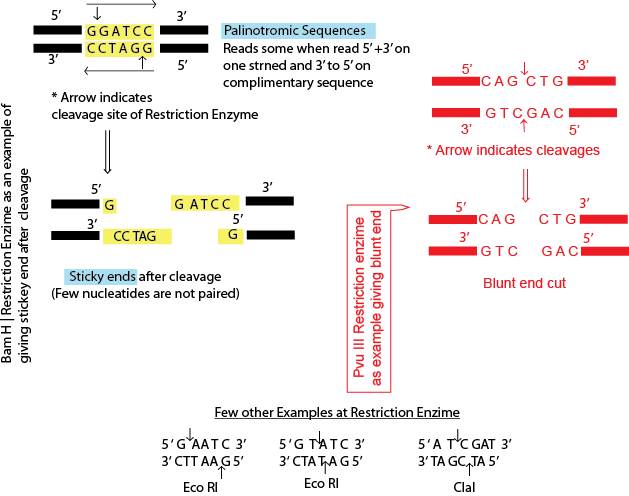
Figure 2: Cleavage sites of restriction endonucleases
|